Zeneth is Lhasa’s expert, knowledge-based in silico software for the prediction of forced degradation pathways of organic active pharmaceutical ingredients (APIs).
Zeneth can provide knowledge and mechanistic understanding of the potential chemical degradation pathways of APIs under defined environmental conditions; those typically used in forced degradation studies (temperature, pH, water, oxygen, peroxide, light, metal ions, radical initiator). Excipients (and their known impurities) can also be evaluated against the API to assess API:excipient interactions, using Zeneth’s built-in excipient database. To this end, the program is primarily employed within the pharmaceutical industry to understand and account for any potential degradation problems during API development. Zeneth also plays an important role in aiding compatibility studies for generics companies.
Zeneth works by considering the chemical structure of a given API and assessing it against the environmental conditions selected by the user using a collection of degradation patterns which are held in a knowledge base. If any part of the API matches a degradation pattern, and the relevant environmental condition is triggered, a degradant structure is generated, see Figure 1.
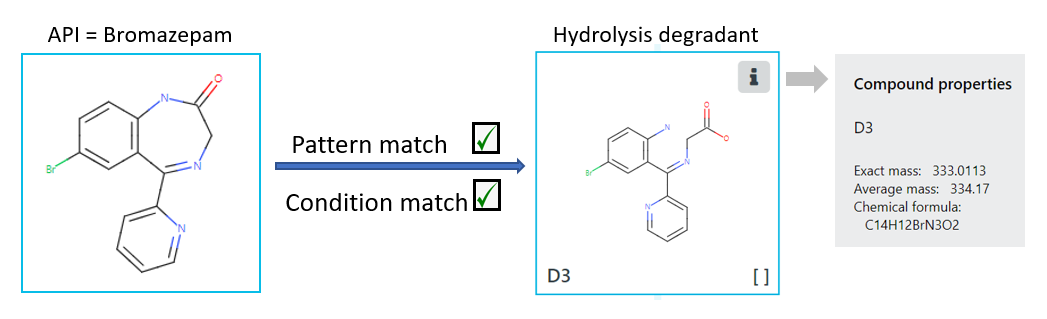 |
Figure 1: Zeneth predicts a hydrolysis degradant (D3) of the API Bromazepam, due to a pattern and condition match in the knowledge base. As well as the structure, the software also generates chemical properties for each degradant, namely exact mass, average mass and chemical formula.
This happens continuously and exhaustively until the program has assessed all its degradation patterns. A score, ranging from 0-1000 (where 0 is impossible and 1000 is certain) will be calculated for each degradant as it is generated. These predicted degradants are then displayed in a tree-like fashion of descending generations of degradants, see Figure 2.
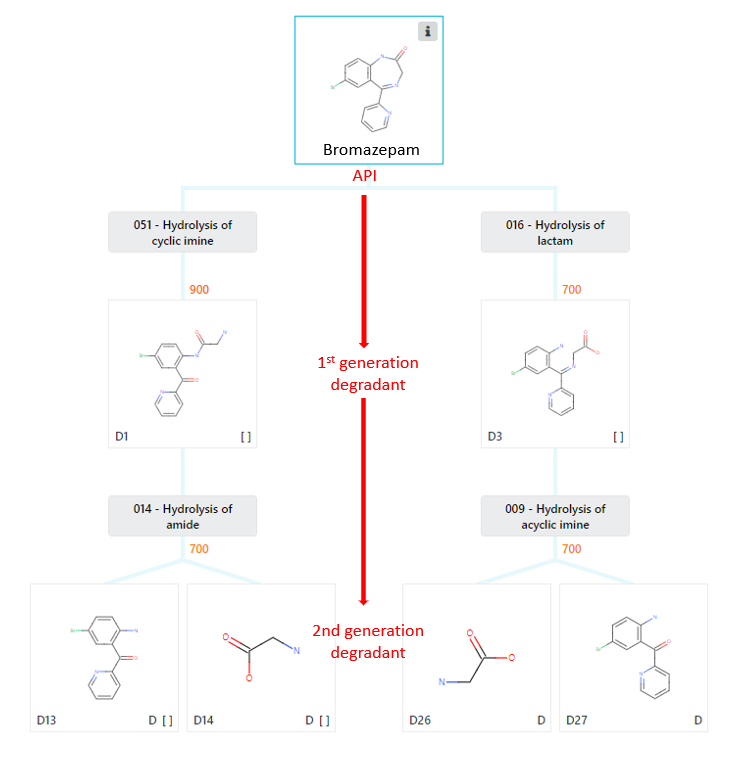 |
Figure 2: Prediction tree showing two generations of hydrolysis degradants for the API bromazepam. These degradants are predicted with scores of either 700 or 900.
To aid analysis, these trees can be filtered to expose specific pathways of interest and searched to locate specific degradants using variables such as exact mass, chemical formula, or formula loss. Intermediate structures are also predicted as part of the degradation pathway and can be searched based on type (observable, tautomer, mechanistic, radical), see Figure 3.
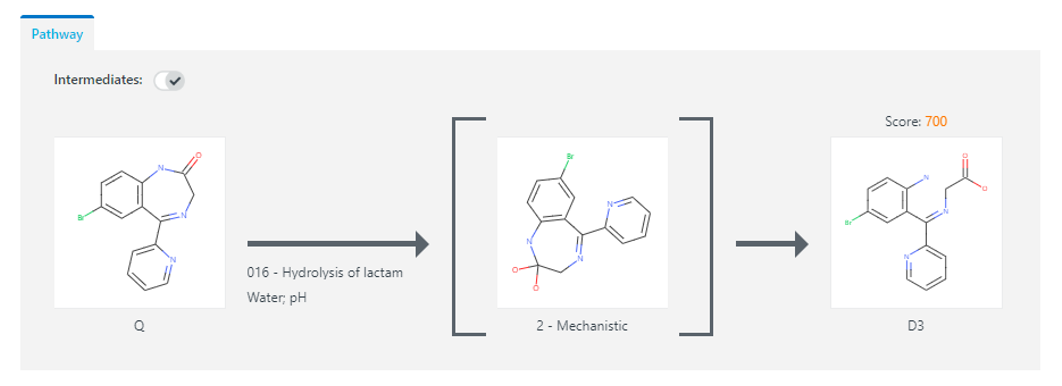 |
Figure 3: A hydrolysis degradation pathway for the API Bromazepam. Environmental conditions as well as a mechanistic intermediate are outlined as part of the route to generate the degradant (D3)
A new feature within the software allows a user to create a spider diagram using the prediction tree. This diagram provides a fast, easy and effective way to focus on degradation pathways of interest, generating a visual representation of this. These diagrams can be saved out of the program as .svg files for upload into other programs (without any compromise to image quality) where further editing can take place (if required). The files are also automatically saved in the software so this information can be revisited at any time.
Upon entering the spider diagram mode, a user is directed to select pathways of interest by highlighting specific degradants, see Figure 4.
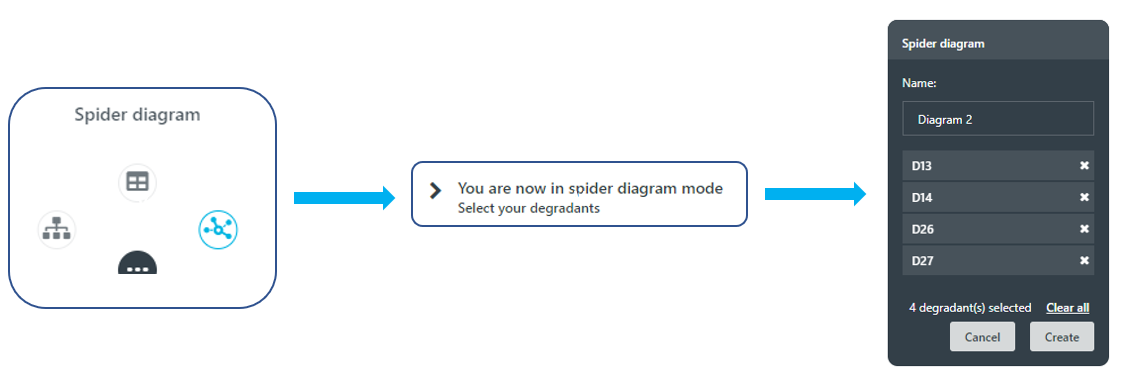 |
Figure 4: Set-up steps to generate a custom-built spider diagram within the Zeneth application.
The software will then create the diagram using the query compound as the focal point, pulling through meta data for each pathway of interest namely the likelihood score and condition triggers, see Figure 5.
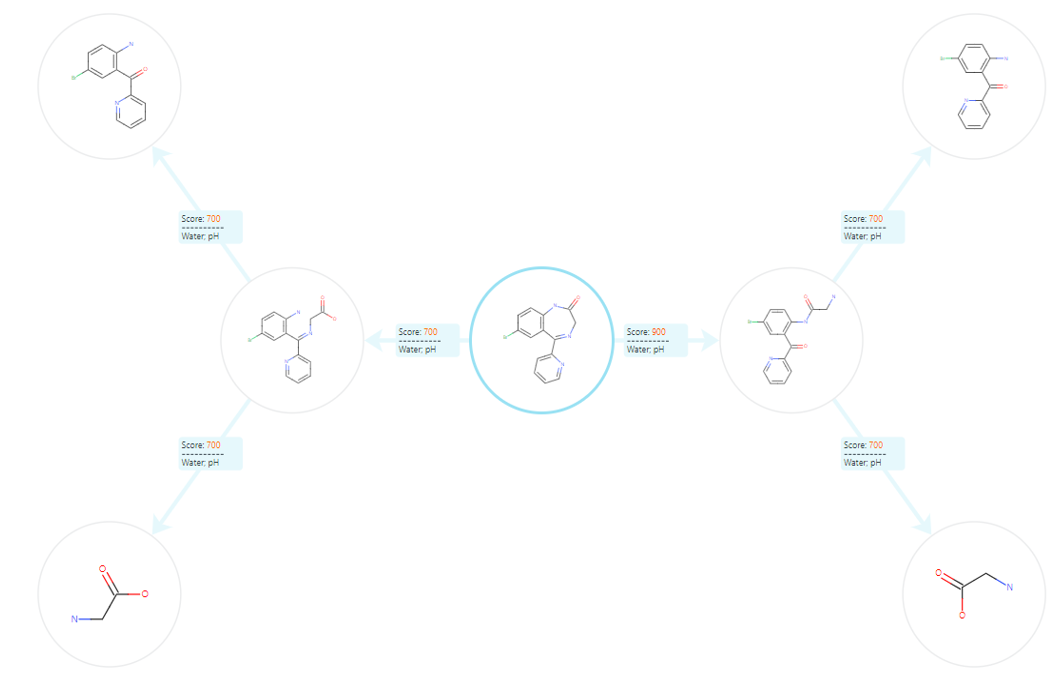 |
Figure 5: Custom generated spider diagram showing degradation pathways of interest for the API Bromazepam. Likelihood scores and condition triggers are outlined for each degradant.
Going forward, we are planning to further develop the functionality of these spider diagrams; adding more features to improve useability and flexibility, thereby increasing value to the user. Additions may include the ability to draw multiple spider diagrams within a single prediction, the inclusion of intermediates for a given degradation pathway and a ‘free-text’ box to allow a user to further customise their diagram.
____________
Did you enjoy this blog? Please let us know.
What else would you like us to write about? Please let us know.



